Background
Protein's 3D structure can be predicted by AlphaFold, an AI system developed by DeepMind with very high accuracy. Although its training set contains a low number of transmembrane structures, because of the structure determination problems of this type of protein, the predicted transmembrane protein structures seem to be also accurate. We use 'seem to be' here, since there are not enough transmembrane protein structures (that are not seen by AlphaFold during the training), to accurately evaluate the accuracy of AlphaFold method on transmembrane proteins.
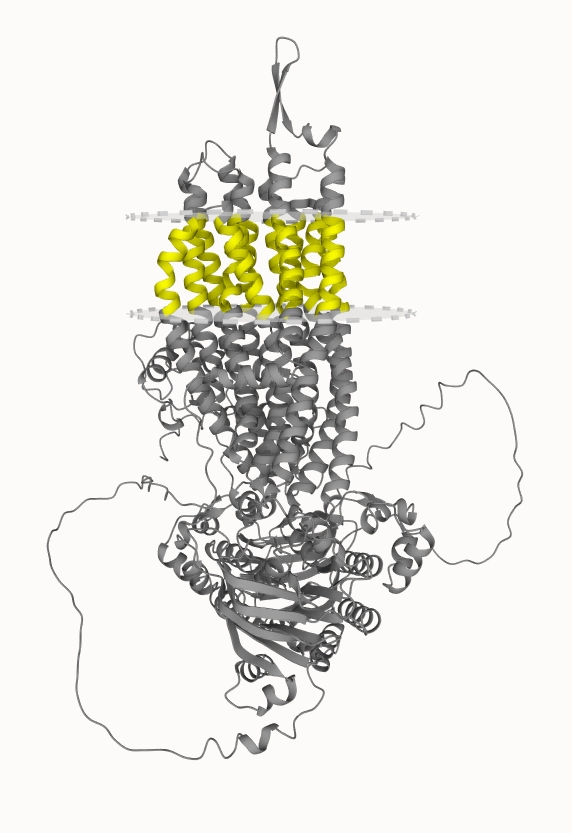
Moreover, similar to the determined 3D structures of transmembrane proteins, a vital component is missing from the structure that is the double lipid membrane layer.
In TmAlphaFold database the relative position of the transmembrane protein structure to the membrane layer is calculated by TMDET algorithm, and according to this calculation an evaluation of the predicted structure is also provided.
TmAlphaFold database contains all alpha-helical transmembrane protein orientation in the membrane deposited so far in AlphaFold database at EBI, and we would like to update its content following the growth of AlphaFold database.
By using TmAlphaFold database you accept the Privacy Notice in compliance with Europe’s new General Data Protection Regulation (GDPR) that applies since 25 May 2018. Read the Privacy Notice