Usage of TmAlphaFold database site
Search modes
 The search bar at the top of the page can be used to search for transmembrane (TM) protein structures. Users can search for gene name, UniProt Accession, UniProt ID or species. After searching, results will be listed, along with basic information about the search hits.
The search bar at the top of the page can be used to search for transmembrane (TM) protein structures. Users can search for gene name, UniProt Accession, UniProt ID or species. After searching, results will be listed, along with basic information about the search hits.
Search results
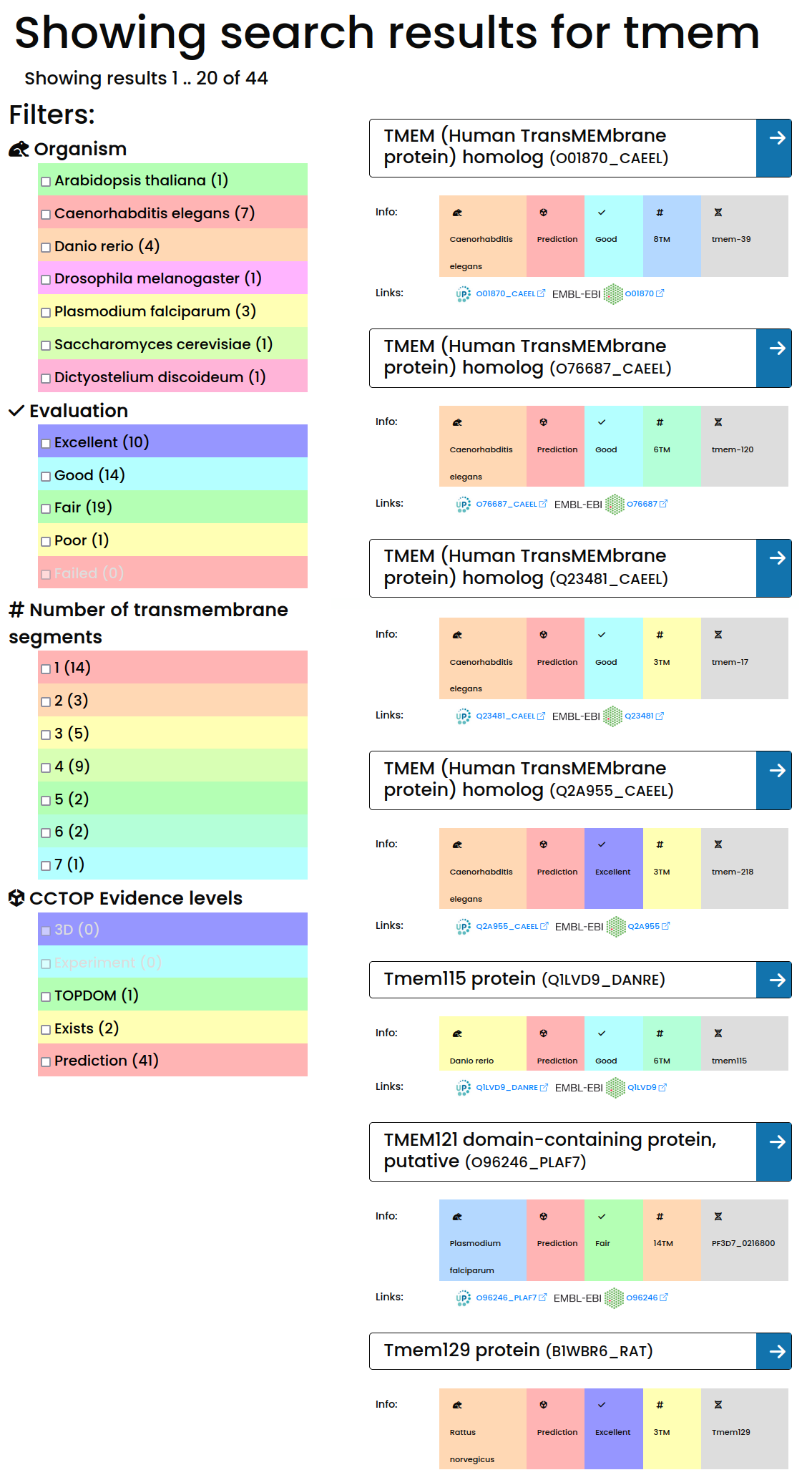 The result page lists all hits from the search term. We display the most important information about the listed proteins, such as protein name, gene name, organism, UniProt AC, together with the AlphaFold Database and PDB-EBI hyperlinks.
Results can be further filtered based on different settings (such as organism, quality of the structure, topology prediction evidence and number of transmembrane helices), using the navigation bar on the left side.
The result page lists all hits from the search term. We display the most important information about the listed proteins, such as protein name, gene name, organism, UniProt AC, together with the AlphaFold Database and PDB-EBI hyperlinks.
Results can be further filtered based on different settings (such as organism, quality of the structure, topology prediction evidence and number of transmembrane helices), using the navigation bar on the left side.
Protein page
After clicking on a protein from the search results, the user is navigated to the protein page, where five panels are available:


-
Information panel
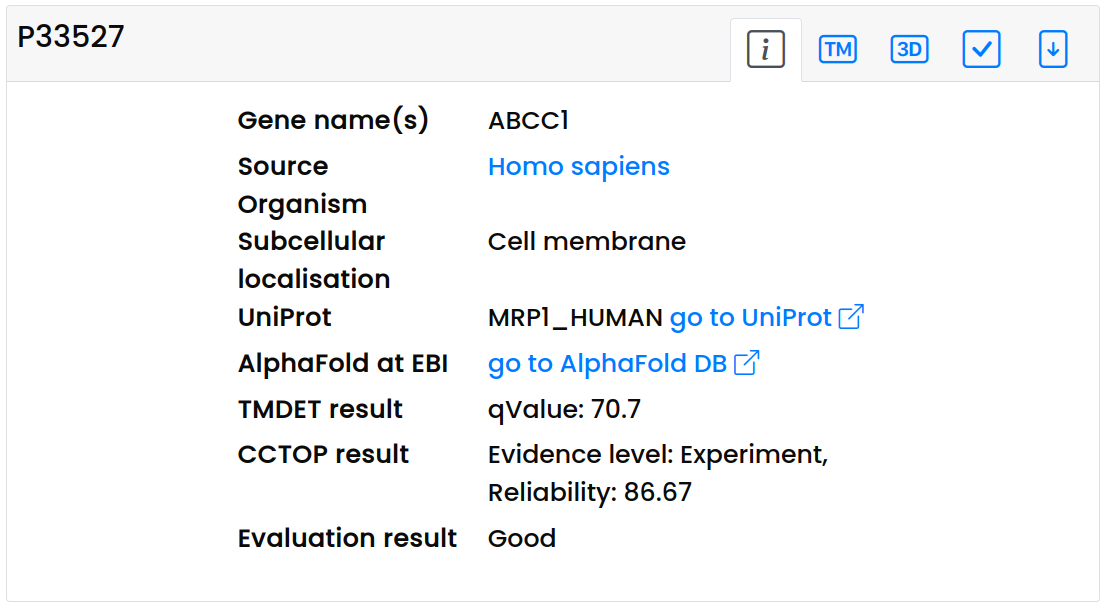
- On the information panel, general information about the protein is available: protein name, gene name, organism, subcellular localization (from UniProt) and a hyperlink to the AlphaFold database. Furthermore, we display the Q value determined by TMDET (that is, how accurate the membrane plane detection by TMDET), and the evidence and reliability levels from CCTOP.
-
Topography panel
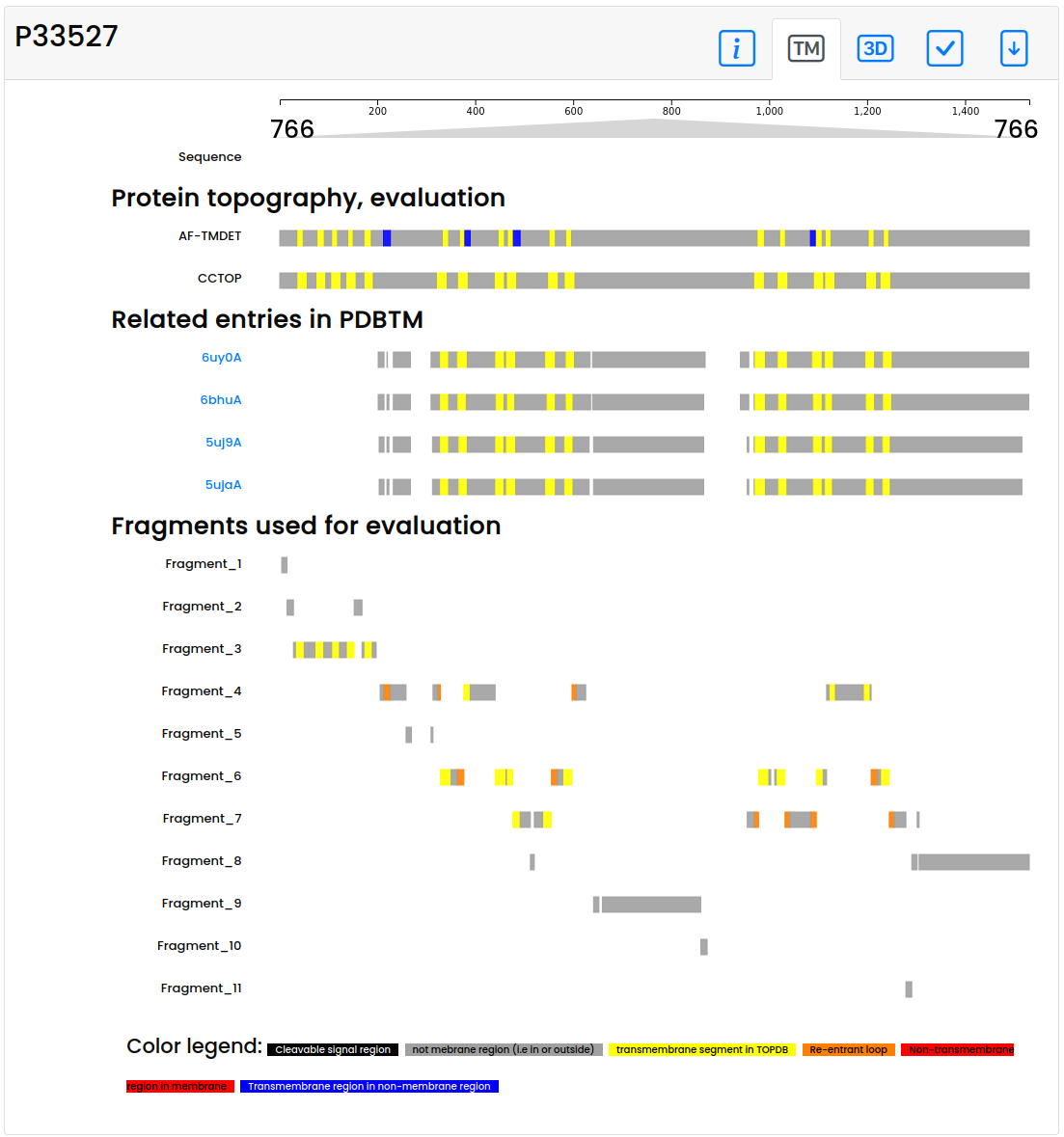
- On the topography panel, the proposed topography of the protein is displayed, considering the TMDET algorithm result on the AlphaFold structure. TM helices that seem to be correct are highlighted with yellow color. Segments in the membrane plane that are not supposed to be there are marked with red (these errors can raise from Short helix, Masked, Masked cross, Domain, Domain cross). In contrast, segments expected to harbor the membrane, yet placed outside the membrane plane are blue. For a quick comparison, the CCTOP topology prediction and related structures from PDBTM are also shown, where yellow lines highlight predicted/detected TM segments. Last, but not least fragments and their membrane segments are also displayed. Note that this tab is only visible if the membrane plane could be reconstructed (thus on Failed structures it may be disabled).
-
3D structure panel
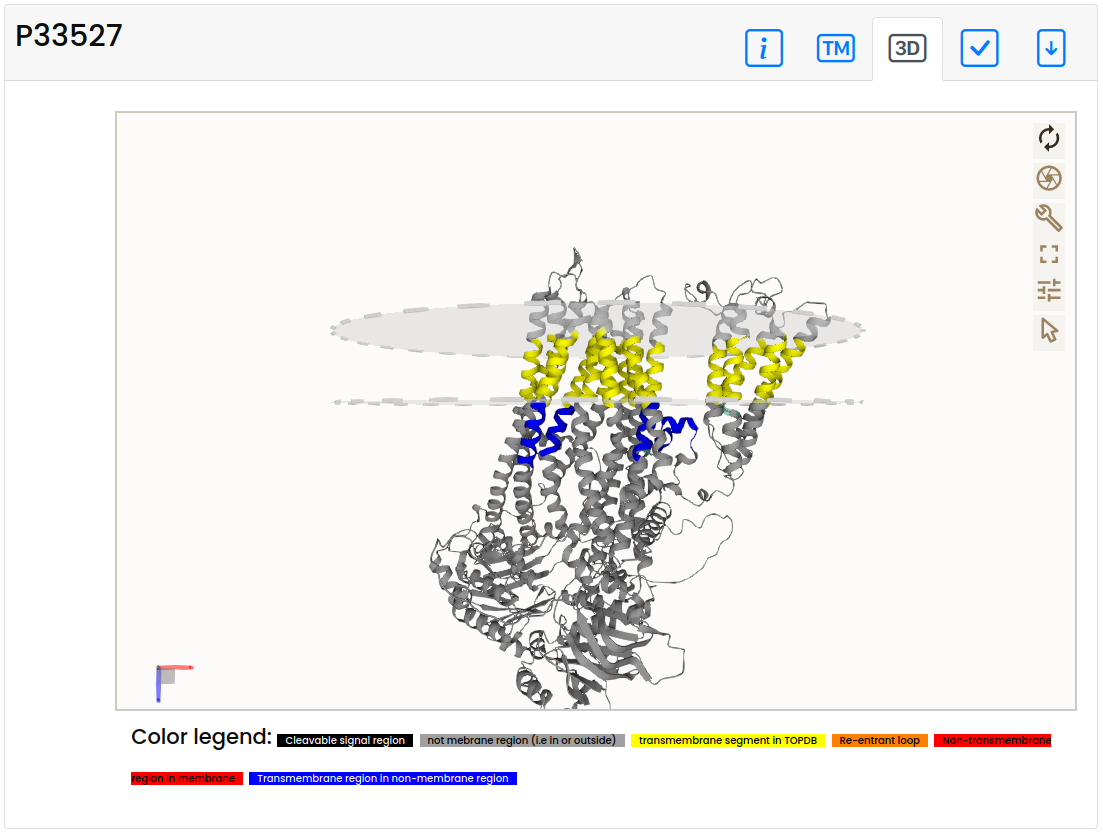
- On the 3D structure panel, the AlphaFold structure is visible with the reconstructed membrane plane. Coloring is the same as described in the topography panel. Note that this tab is only visible if the membrane plane could be reconstructed (thus on Failed structures it may be disabled).
-
Evaluation panel
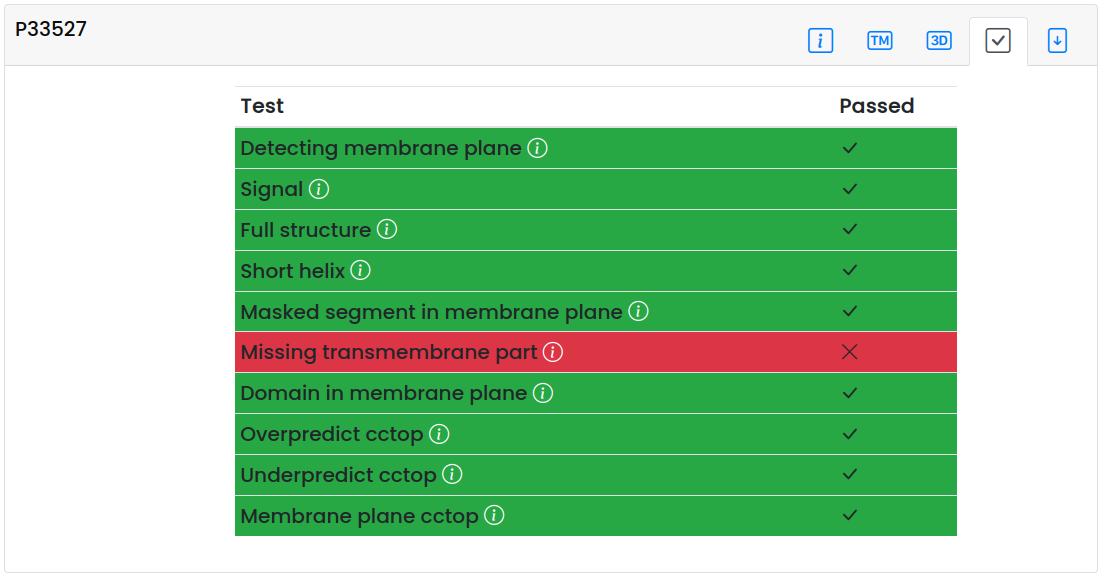
- On the evaluation panel, the result of the quality checks are visible. For detailed information what is the meaning of each flag, hover mouse over them for quick help or visit the Methodology page.
-
Download panel
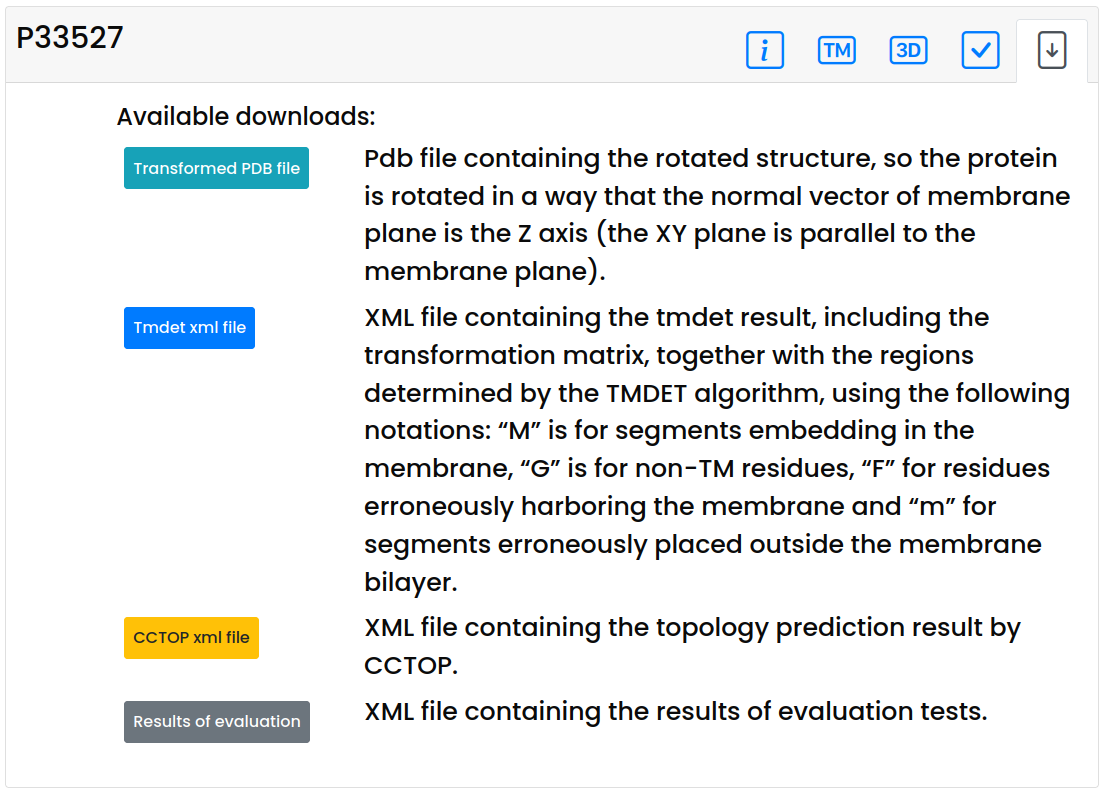
-
On the download panel users can download the results in various formats:
- The AF.trpdb.gz file contains the rotated structure, so the protein is rotated in a way that the normal vector of membrane plane is the Z axis (the XY plane is parallel to the membrane plane)
- The AF.xml file contains the tmdet result, including the transformation matrix, together with the regions determined by the TMDET algorithm, using the following notations: “M” is for segments embedding in the membrane, “G” is for non-TM residues, “F” for residues erroneously harboring the membrane and “m” for segments erroneously placed outside the membrane bilayer.
- The cctop.xml file contains the topology prediction result by CCTOP.
- The AFTM.xml file contains the results of evaluation tests.
Statistics page
There are several pre-calculated statistical data on the Statistics page (number of entries
by evaluation categories,
by number of transmembrane segments,
by CCTOP prediction results evidences and
by species), but user can generate any other chart by clicking the button.
First and second categories for creating chart can be selected by clicking the appropriate icon on the right side panel and the element of the selected categories can be chosen under the "Select elements" tab. After creating the chart user can "freeze" it (i.e. it cannot be modified further) by clicking the button, or user can delete it by clicking the button.
User can add more than one chart to the page by repeatedly clicking the button, or can be freeze the whole page by clicking the at the bottom right side of the page. The url of the generated page is unique and it can be decoded by the server, therefore it can be easily shared or bookmarked for later usage.
First and second categories for creating chart can be selected by clicking the appropriate icon on the right side panel and the element of the selected categories can be chosen under the "Select elements" tab. After creating the chart user can "freeze" it (i.e. it cannot be modified further) by clicking the button, or user can delete it by clicking the button.
User can add more than one chart to the page by repeatedly clicking the button, or can be freeze the whole page by clicking the at the bottom right side of the page. The url of the generated page is unique and it can be decoded by the server, therefore it can be easily shared or bookmarked for later usage.
Download page
Membrane orientation, CCTOP prediction results, rotated structure in pdb format and the evaluation can be downloaded for each entry using the download () tab at the entry viewer page. We also prepared sets to bulk download structures separated by organisms, which can be downloaded from download page.
By using TmAlphaFold database you accept the Privacy Notice in compliance with Europe’s new General Data Protection Regulation (GDPR) that applies since 25 May 2018. Read the Privacy Notice